Learning
Bayesian network using EM algorithm from Incomplete Data
Problem Statement
We often encounter missing values or hidden variables in learning Bayesian networks(BNs) from medical data. However, most existing state of the art learning algorithms are not able to learn structure from incomplete data. Hence, I propose to use EM algorithm to learn BNs from incomplete data.
Methods
There are two major components for Bayesian Network learning:
● to Implement EM algorithm for parameters learning ,i.e. conditional probability table
● to learn structures using Bayesian Information Criterion (BIC)
Data
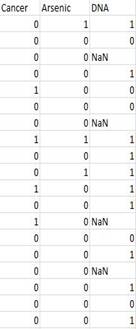
The data I used came from Dartmouth medical school. The
table below shows a portion of the data I used for this analysis. This date set
consists of 3 variables and 1113 observations. About 20% of values in DNA are
missing. These three variables are arsenic exposure,Xrcc3 DNA genotype, bladder
cancer. According to a recent study from Dartmouth Medical school, there is
evidence of gene-environment interaction between XRCC3 variant genotype and
high toenail arsenic levels on bladder cancer risk (Andrew 2009a).
Brief Introduction to Bayeisan
Network
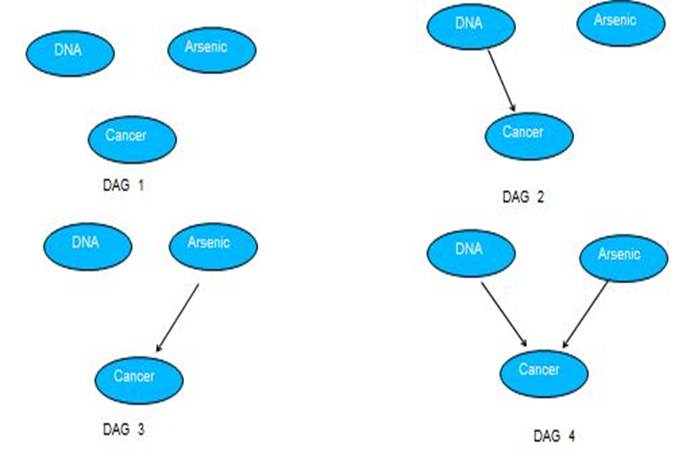
The Bayesian network below is a suggested one based on Dr. Andrew's study, which illustrates interactions among those three variables. In this DAG, arsenic exposure and DNA are found to influence cancer. There conditional probability tables associated with each node are also given.
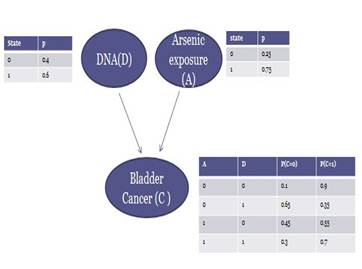
A typical BN consists a directed acyclic graph(DAG) and the conditional
probability tables. The edges represent causal relationship between variables.
For a particular BN, the joint probability of variables is shown in the
following function:
![]()
Parameters
learning from missing data
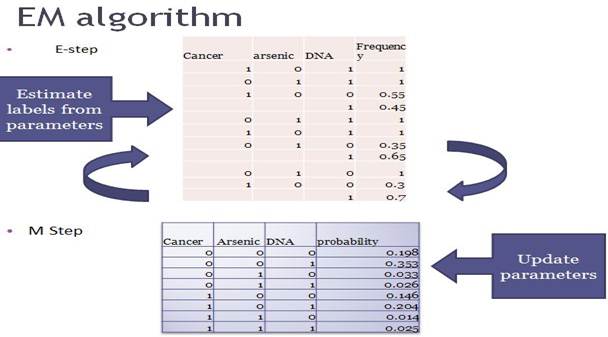
In the E-step, the algorithm estimates the labels of missing data from parameters. For each observation with missing values, the algorithm assign frequency to them based on the conditional probability tables. After each E-step, the date set are updated, from which we can future derives new parameters in the M step. The whole process repeats until it reaches convergence, i.e. when the log-likelihood of parameters stop changing. The log-likelihood of the entire data is shown in the function below:
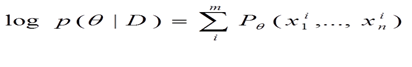
Structure
Learning
Once parameters are learned, we can search through all possible structures. According to the prior knowledge, the relationship between DNA and Arsenic exposure are independent, hence only four possible structures or DAGs are listed below. The algorithm searches through the space and find the structure that optimally describe the data set based on penalized likelihood score, which is Bayesian information criterion(BIC) in this model . BIC score is described in the following function , while L is the maximized value of the likelihood function for the estimated model; k is the number of parameters; m is sample size. The structure with lowest BIC score is the optimal structure.
![]()
Results
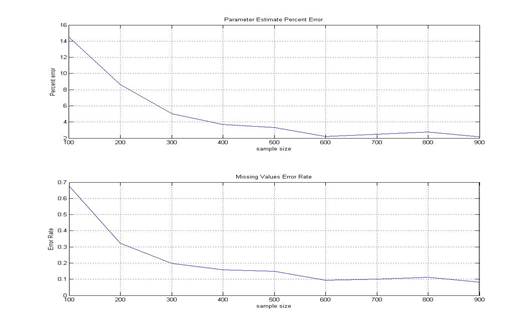
The EM algorithm estimates parameters more accurately as the sample size increases. From the percent error graph, percent error is close to 2.16% when the sample size is 900.
Percent error= ![]() Error
rate for missing values also drops as sample size increases. When the sample
size is 900 the error rate is 0.0812.
Error
rate for missing values also drops as sample size increases. When the sample
size is 900 the error rate is 0.0812.
,
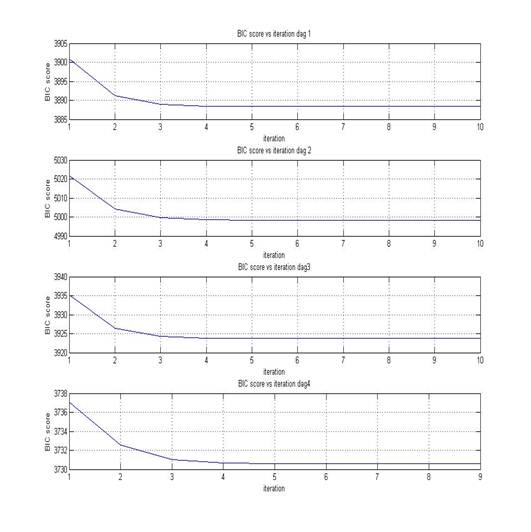
The graph below shows BIC scores versus EM iteration. We can see that DAG 4 has the lowest BIC score. Thus DAG 4 is the best model learned by the algorithm, which is consistent with structure proposed in the introduction
Conclusion
I have accomplished what I have proposed in the project proposal and the algorithm satisfactorily estimates parameters using EM method and the BN learned from this algorithm is consistent with the Dr. Andrew's study. However, the current model can only be applied to data with small size of variables. Besides, since we assume that there are no hidden variables apart from DNA, arsenic and cancer, the BN learned in this model might be different with the inclusion of other caner risk factors such as gender, ages and smoking. Future works would be to develop models that can handle more variables and more complicate BNs.
References:
Andrew, A.S., Mason, R.A., Kelsey, K.T., Schned, A.R., Marsit, C.J., Nelson, H.H., Karagas, M.R., DNA repair genotype interacts with arsenic exposure to increase bladder cancer risk. Toxicol. Lett., 187,10-14, 2009a.
Friedman,
N. 1998. The Bayesian structural EM algorithm. In Proceedings of the Fourteenth
Conference on Uncertainty in Artificial Intelligence (UAI-98), Cooper, G. F.
& Moral, S. (eds). Morgan Kaufmann,129-138.
Darwiche,Adnan ., Modeling and Reasoning with Bayesian Networks. Cambridge University Press, 446 pp., Cambridge, UK, 2009.