Structural EM Algorithm for Learning
Bayesian Networks from Incomplete Data
Introduction
A
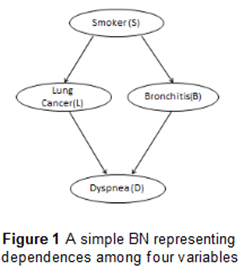
Bayesian network (BN) is a directed acyclic graph (DAG) in which nodes
represent random variables and arrows represent
probabilistic
dependencies between them. Figure 1 shows simple BN representing dependences among four variables.
There
is an edge from S to L because Smoking has a direct influence on the presence
of Lung Cancer. Smoking also
has
a direct influence on the occurrence of Bronchitis. Dyspnea
(shortness-of-breath) may be due to Lung cancer or
Bronchitis
or both of them. BNs can be effective tools for
combining prior knowledge with observational data
to infer model causal relations. Many existing
learning algorithm can recover BNs from complete data. However,
we often encounter missing values or hidden variables
in learning BNs in real-life application.
People may simply
discard the missing values or replace them with
certain values. Either of these approaches may lead to distorted BNs.
In this project, structural Expectation-Maximization
(SEM) (Friedman, 1998) will be implemented to learn BNs from
incomplete data.
Method
Friedman presented a structural EM
algorithm for learning Bayesian network structures in the presence of missing
values
and hidden variables in 1998. The
search over the space of Bayesian networks alternates between two steps: an
optimization for the Bayesian
network parameters conducted by the EM algorithm, and the structural search for
a
better Bayesian network structure
using the hill climbing strategy. At each step, it can either find better
parameters for the current
structure, or selected a new structure. The former case is a standard
"parametric"
EM step, while the latter is a
standard "parametric" EM step.
Data
I
have available a rich genetic epidemiological data set from a large,
population-based, case-control study of bladder
cancer
in New Hampshire. These data include over 1477 SNPs in cancer-related genes,
detailed smoking assessment,
gender,
age, as well as other risk factors including arsenic exposure. (Karagas et,al,
1998, Karagas et al. 2004).
Milestone Goals
1.
Implement
EM algorithm to learn parameters
04/10-04/24
2. Implement Hill
climbing to learn BNs
04/25-05/10
3. Write up milestone
report
05-06-05-07
4. Testing and polishing
results
05/11-05/20
5.
Write
up final report 05/20-05/25
Reference
Karagas, M.R.,
Tosteson, T.D., Blum, J., Morris, J.S., Baron, J.A. and Klaue, B., Design of an
epidemiologic study
of drinking water arsenic exposure and
skin and bladder cancer risk in a U.S. population. Environ. Health
Perspect., 106(Suppl 4), 1047-1050, 1998.
Karagas, M.R.,
Tosteson, T.D., Morris, J.S., Demidenko, E., Mott, L.A., Heaney, J. and Schned,
A., Incidence of
transitional cell carcinoma of the bladder
and arsenic exposure in New Hampshire. Cancer
Causes Control,
15, 465-472, 2004.
Friedman, N. 1998.
The Bayesian structural EM algorithm. In Proceedings of the Fourteenth
Conference on
Uncertainty in Artificial Intelligence
(UAI-98), Cooper, G. F. & Moral, S. (eds). Morgan Kaufmann,129-138.
Guo, Y.-Y., Wong,
M.-L. & Cai, Z.-H. 2006. A novel hybrid evolutionary algorithm for learning
Bayesian networks
from incomplete data. In Proceedings of the
IEEE Congress on Evolutionary Computation (CEC 2006),916-923.